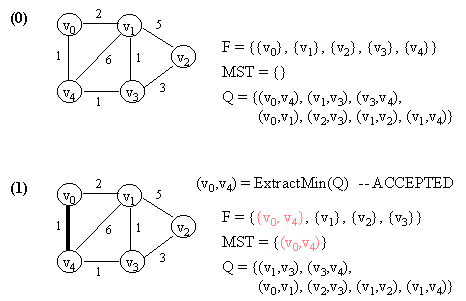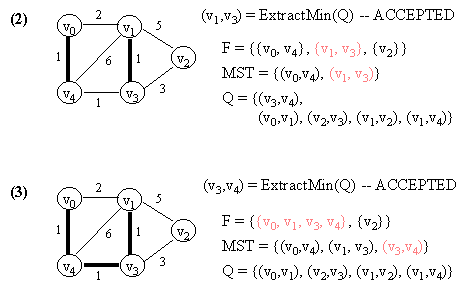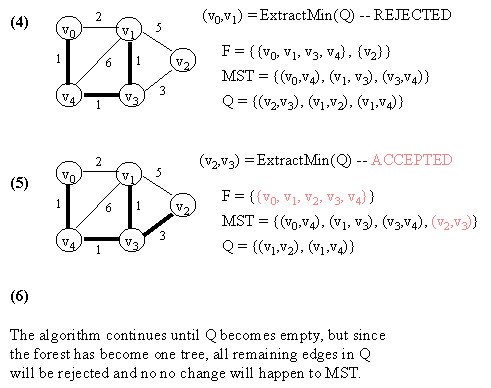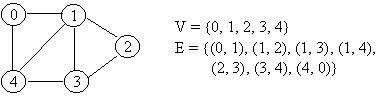

- (v0, e1, v1, e2, v2,..,vn-1, en, vn) -- alternating vertices and edges
- (v0, v1, v2,..,vn-1, vn) -- vertices only
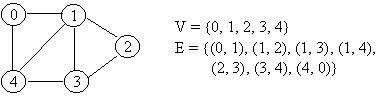


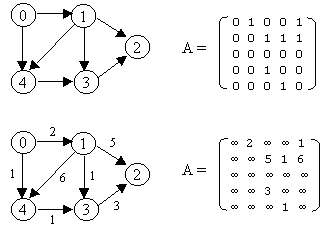
Graphs G1 and G2 are isomorphic if there is a one-to-one, onto function f from the vertices of G1 to the vertices of G2 and a one-to-one , onto function g from the edges of G1 to the edges of G2 , so that an edge e is incident on v and w in G1 if and only if the edge g(e) is incident on f(v) and f(w) in G2. The pair of functions f and g is called an isomorphism of G1 onto G2.
Theorem : Graphs G1 and G2 are isomorphic if and only if for some ordering of their vertices, their adjacency matrices are equal.
Corollary : For simple graphs G1 and G2, they are isomorphic if and only if there is a one-to-one, onto function f from the vertex set of G1 to the vertex set of G2 satisfying the following: Vertices v and w are adjacent in G1 if and only if the vertices f(v) and f(w) are adjacent in G2.
A graph is planar if it can be drawn in the plane without its edges crossing.
Euler's Formula for Planar Graph
In any simple, connected, planar graph G , we always have e £ 3v - 6
K5 and K3,3 are not planar.
Kuratowski's Theorem : A graph G is planar if and only if G does not contain a subgraph homeomorphic to K5 or K3,3 .
Input : A connected weighted graph with positive weighted, vertice a
and z
Output : L(z) , the length of a shortest path from a to
z .
1. procedure dijkstra(w,a,z,L)
2. L(a) := 0
3. for all vertices x ¹ a do
4. L(x) := ¥
5. T := set of all vertices
6. // T is the set of vertices whose shortest distance from a has
7. // not been found
8. while z Î T do
9. begin
10. choose v Î T with minimum L(v)
11. T := T - {v}
12. for each x Î T adjacent to v do
13. L(x) := min{ L(x), L(v) + w(v,x)}
14. end
15. end dijkstra
For input consisting of n-vertex, simple, connected, weighted graph,
Dijkstra's Algorithm has worst-case run time Q
(n2) .
A cycle (path) in a graph G that includes all of the edges (as a consequence, also includes all the vertices) is called an Euler cycle (path).
The necessary and sufficient condition for a graph G to have a Euler cycle is
The necessary and sufficient condition for a graph G to have a Euler path is
A cycle in a graph G that contains each vertex in G exactly once, except for the starting and ending vertex that appears twice.
A path in a graph G that contains each vertex in G exactly once is called a Hamiltonian path.
There is no well-known necessary and sufficient condition for the exitence of a hamitonian cycle in a given graph G.
Given a weighted graph G, find a minimum-length Hamiltonain cycle for G.
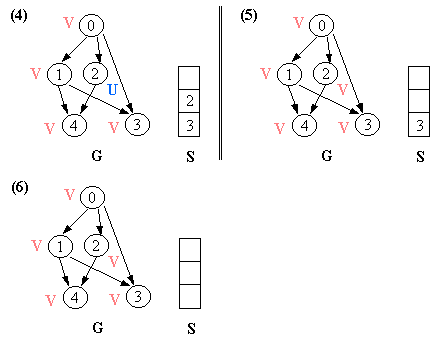
8.1 Depth-First Traversal
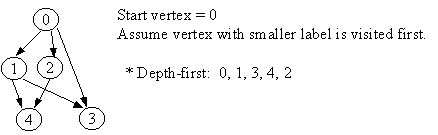
Let G = (V, E) is a graph which is represented by an adjacency matrix Adj. Assume that nodes in a graph record visited/unvisited information.
procedure DEPTH-FIRST (G) 1. Initialize all vertices as "unvisited". 2. Let S be a stack. 3. Push the root on S. 4. While S not empty, do 5. begin 6. Let n <- Pop S. 7 If n is marked as "unvisited", then 8. begin 9. Mark n as "visited", and output n to the terminal. 10. For each vertex v in Adj[n], do 11. If v is marked as "unvisited", then // this test is actually redundant 12. push v on S. 13. end 14. end

8.2 Breadth-First Traversal
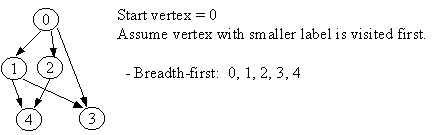
procedure BREADTH-FIRST (G) 1. Initialize all vertices as "unvisited". 2. Let Q be a queue. 3. Enqueue the root on Q. 4. While Q not empty, do 5. begin 6. n <- Dequeue Q. 7. If n is marked as "unvisited", then 8. begin 9. Mark n as "visited", and output n to the terminal. 10. For each vertex v in Adj[n], do 11. If v is marked "unvisited", then 12. enqueue v on Q.13. end 14. end

10.1 Prim's Algorithm
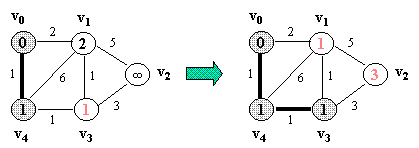
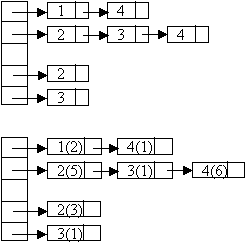
Let G = (V, E) which is represented by an adjacency list Adj. Some support data structures:
PRIM(G, s)
1. Initialize d[s] with 0, P[s] with 0, and
all other d[i] (i!=s) with a positive infinity and
p[i] (i!=s) with 0.
2. Q <- V // initialize Q with all vertices as UNKNOWN
3. while Q not empty do
4. begin
5. u <- ExtractMin(Q) // Q is modified
6. Mark u as KNOWN // Dequeing u is the same as marking it as KNOWN
7. for each vertex v in Adj[u] do
8. begin
9. if v is UNKNOWN and d[v] > weight(u, v), then do
10. begin
11. d[v] = weight(u, v) // update with shorter weight
12. p[v] = u // update v's parent as v
13. end
14. end
15. end
Example (NOTE: v0 is the source vertex, and d[i] for each vertex i is also indicated in its circle):
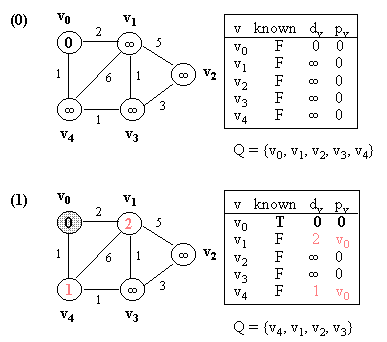
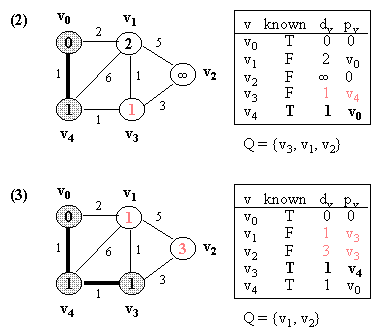
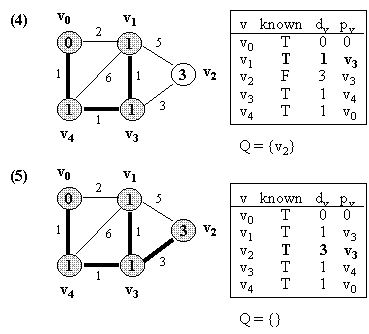
10.2 Kruskal's Algorithm
Let G = (V, E) which is represented by an adjacency list Adj. Some support data structures:
KRUSKAL(G)
1. Let F be a set of singleton set of all vertices in G.
2. MST <- {}
3. Q <- E
4. while Q not empty do
5. (u, v) <- ExtractMin(Q) // Q is modified
6. if FIND-SET(u) != FIND-SET(v) then // FIND-SET(i) returns the set in F
// which vertex i belongs to.
// This effectively does cycle check.
// If ACCEPTED,
7. begin
8. merge(FIND-SET(u),FIND-SET(v)) in F
9. MST <- MST Union {(u, v)}
10. end
11. return MST.
NOTE: In the figure below, a number in a vertex indicates the vertex number (NOT any kind of value).